Discover where custom oligos, produced on-demand by Enzymatic DNA Synthesis, can take you.
EDS produces primers that meet the strict quality requirements for SYBR Green-based qPCR
Amplification of DNA or cDNA in the presence of an intercalating dye enables absolute or relative quantification of accumulating reaction products, as well as the amplification template. SYBR Green-based qPCR is a cost-effective and high-throughput method for gene expression studies, genotyping, food safety or pathogen testing, among many other applications. Specificity is a key performance parameter in qPCR, as intercalating dyes impact the efficiency of DNA polymerases, and qPCR readouts (amplification and melt curves) only provide indirect evidence that the reaction product contains the target of interest. Careful primer design, optimized qPCR chemistry and appropriate experimental design (e.g., the inclusion of positive and no-template controls) are critical to ensure low false-positive rates. Primer quality is important to minimize non-specific priming and primer-dimer formation, which decrease amplification efficiency and may skew results.
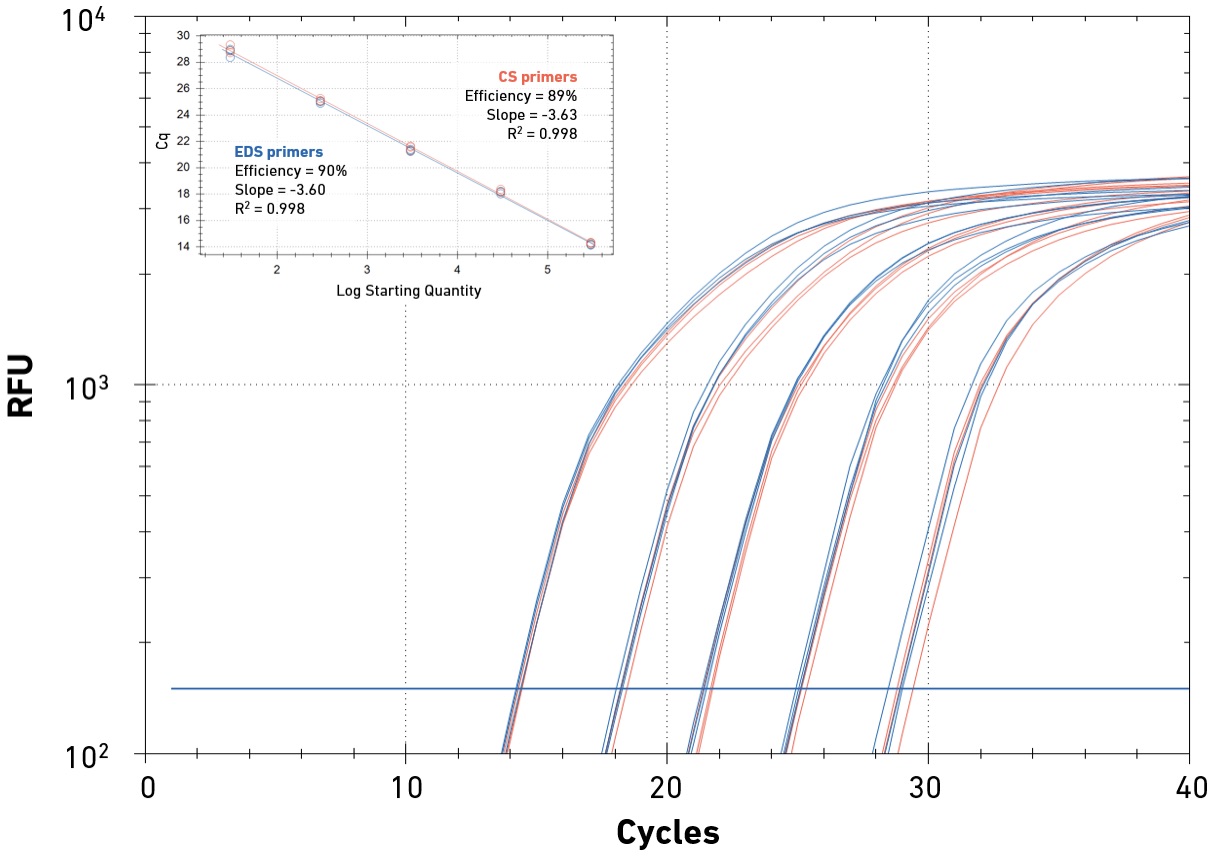
EDS primers support high specificity and reaction efficiency
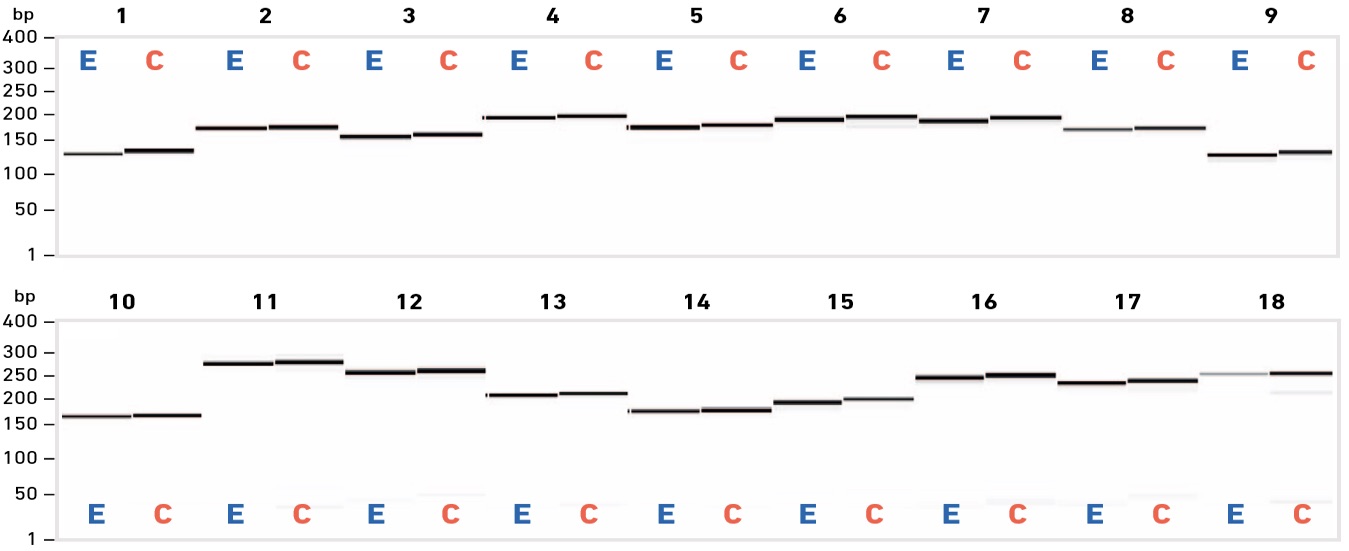
End-point PCR steps are routinely included in laboratory protocols to (i) confirm the presence of one or more specific DNA fragment(s), and/or (ii) generate a sufficient quantity of material for downstream analysis (e.g. sequencing) or manipulation (e.g. mutagenesis, cloning). Irrespective of the application, high primer quality is needed to achieve the sensitivity, specificity and amplification efficiency required for downstream success.
The SYNTAX System offers benefits for high-throughput sgRNA production by in vitro transcription (IVT). Guide RNAs can be produced, evaluated, and optimized in rapid cycles of iteration, each only requiring a few days.
The past decade has seen rapid and widespread adoption of CRISPR/Cas9-based methods for genome editing in biological systems ranging from cultured cells to entire animals. The CRISPR/Cas9 system requires two components: a Cas9 nuclease that cleaves the target sequence, and a single guide RNA (sgRNA) that directs the nuclease to the targeted cleavage site. The sgRNA is composed of two parts: a target-specific CRISPR RNA (crRNA) sequence, and a trans-activating crRNA (tracrRNA) scaffold sequence that is common to all sgRNAs. Successful in vivo gene editing depends on delivery of the Cas9/sgRNA complex to the target genome. In vitro assembly of the Cas9-ribonucleoprotein (RNP) complex is gaining popularity as i) it does not require cloning, ii) allows for high-throughput production of fully functional Cas9-RNP complexes that are active upon delivery, and iii) are quickly degraded, thereby reducing the potential for off-target effects. In preliminary proof-of-concept experiments, we have observed no difference in the efficiency of sgRNA IVT production nor the nuclease activity of crRNA oligos produced by EDS vs. those sourced from a commercial supplier.
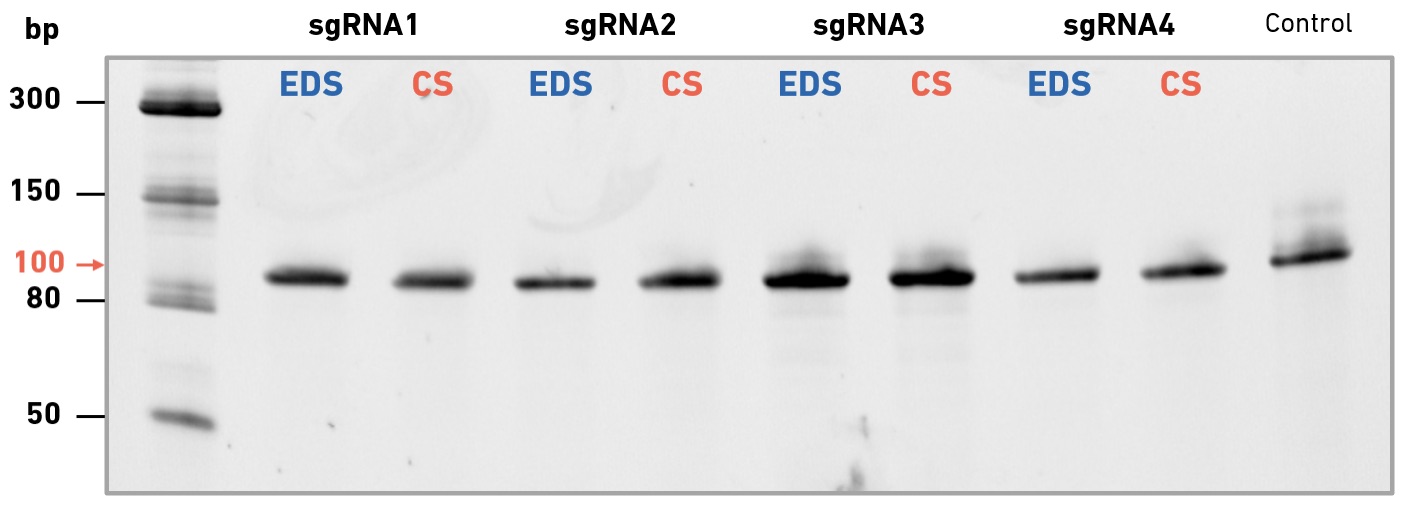
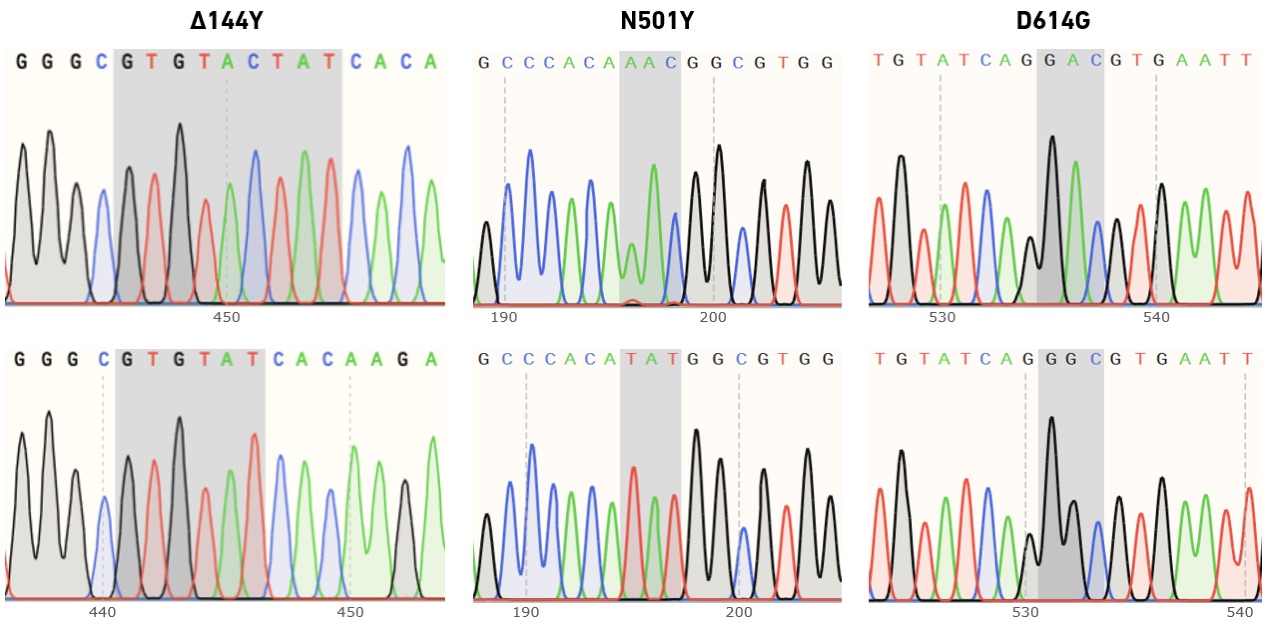
EDS primers enable high success rates in multi-site mutagenesis, and produce high-quality Sanger sequencing data
In collaboration with Felix Rey, Structural Virology Lab at the Institut Pasteur, we have evaluated the performance of enzymatically synthesized oligos in site-directed mutagenesis, followed by variant confirmation using Sanger sequencing. The Rey lab studies viruses of global public health and/or veterinary concern. During the COVID-19 pandemic they have focused on SARS-CoV-2 variants of concern—particularly those with diverse mutations in the spike (S) protein, which may be associated with increased transmissibility or immune escape. The SYNTAX System was used to produce (i) primers for use with the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies), (ii) sequencing primers for the confirmation of individual mutations in picked clones, and (iii) primers for tiled sequencing of the entire S gene.
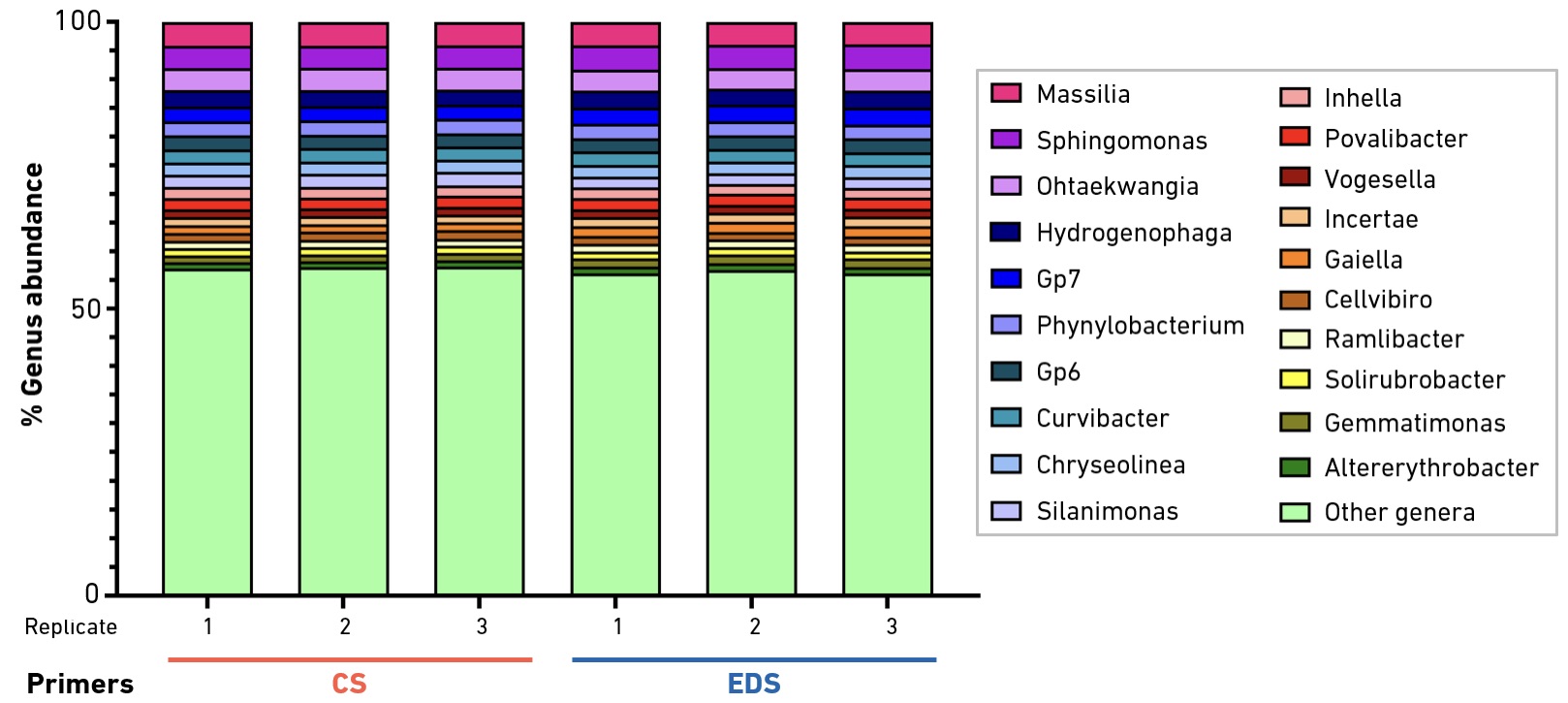
Primers produced by EDS are suitable for a wide range of amplicon sequencing applications. In metagenomic studies, the SYNTAX System offers unique support for deeper interrogation at a rapid pace.
16S amplicon sequencing offers a high-throughput and cost-effective method for the identification and relative quantification of the archaea and bacteria present in complex microbial populations. The method is based on amplification and sequencing of ≥1 of the 9 variable regions of the ~1.6-kb 16S rRNA gene. To evaluate the performance of EDS primers, the well-characterized, degenerate 515F/806R primer pair was synthesized using the SYNTAX System, or ordered from a commercial supplier. Primers were used to generate V4-V5 amplicons from total DNA extracted from a soil sample collected near the Eiffel Tower in Paris, France. Amplification products were converted to libraries for Illumina® sequencing. Bioinformatic analysis confirmed no significant differences in results obtained with EDS vs. commercial primers.
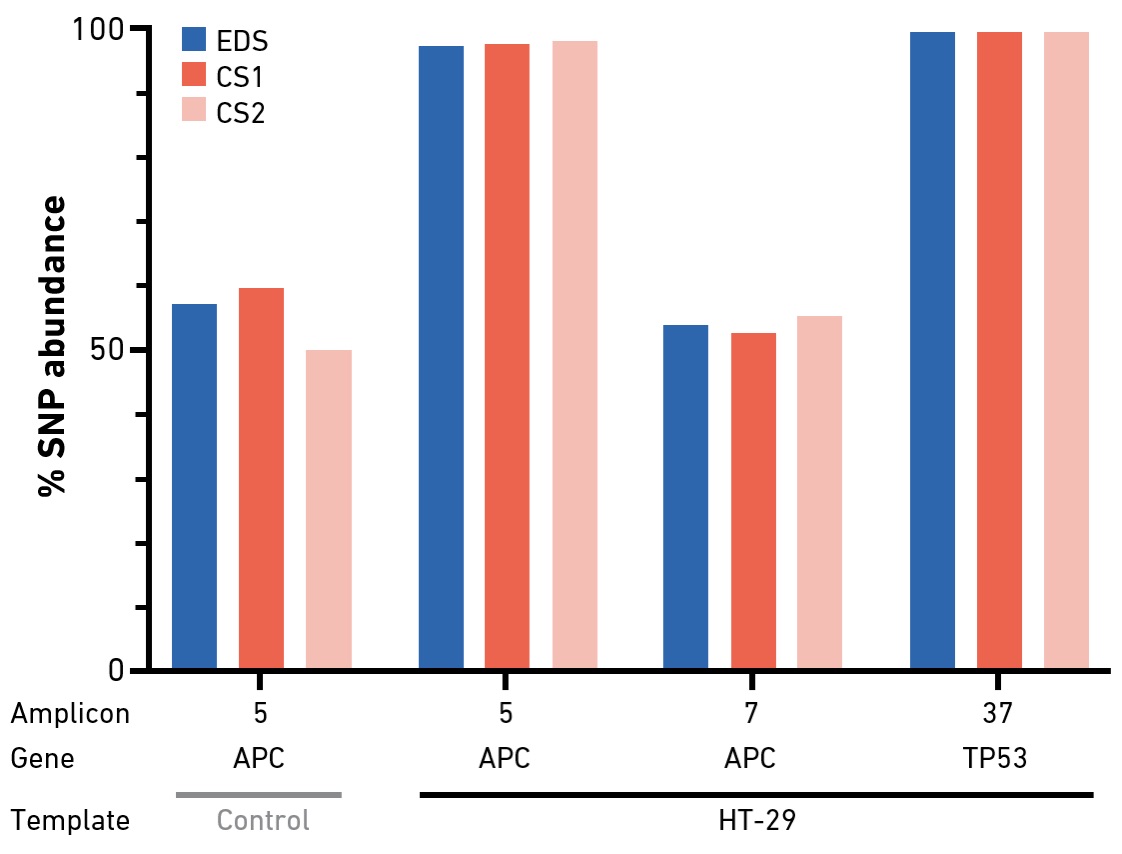
Primers produced with the SYNTAX System are highly suitable for targeted amplicon sequencing. Small panels (up to 48 primer sets per run) can be produced in a standard workday.
Targeted sequencing enables the interrogation of specific genes of interest by NGS. Amplicon-based approaches—in which the regions of interest are amplified by multiplex PCR (in one or multiple pools)—offer streamlined and scalable protocols for library preparation. Despite challenges related to coverage uniformity and duplication rates, deep amplicon sequencing is widely used in cancer research, as input DNA requirements for PCR-based protocols are typically lower than for methods that rely on hybridization capture. In an experiment with a hot spot panel for colorectal cancer genotyping, EDS primers performed comparably or better than primers from two commercial suppliers in terms of overall sequencing data quality, amplicon uniformity and variant calling results. In addition, the system enables on-demand synthesis of primers that may be needed to optimize panel performance (amplicon coverage) or add additional content.
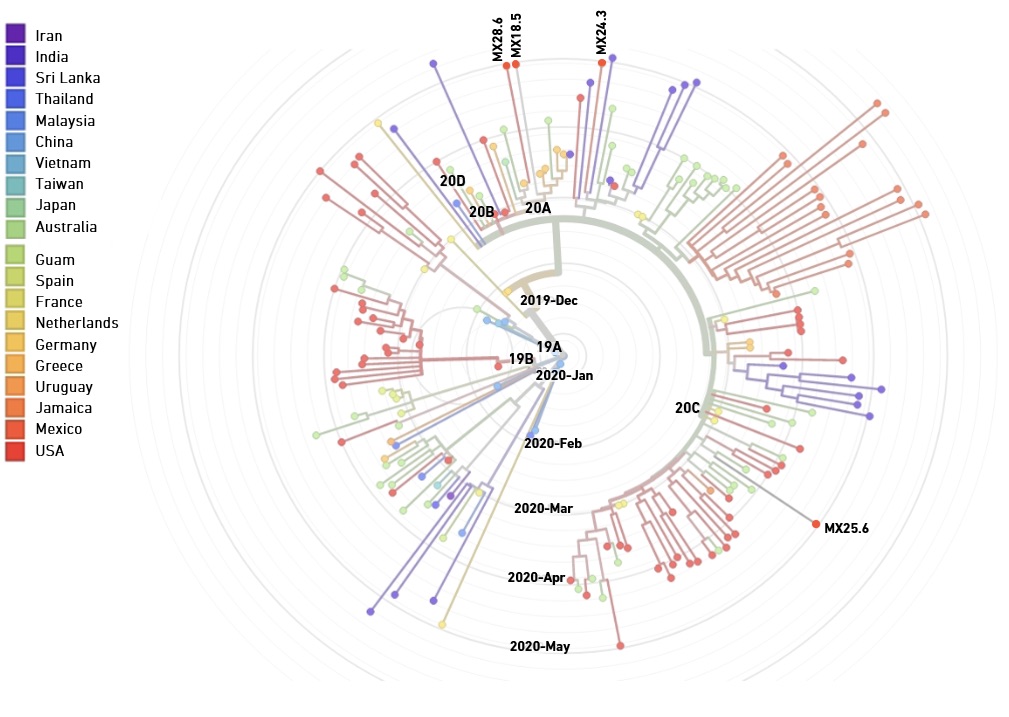
ARTIC V3 primers synthesized by EDS perform comparably to commercial primers with respect to coverage uniformity and variant calling, and support phylogenetic analysis of clinical isolates
In collaboration with The Jackson Laboratory for Genomic Medicine, the University of Zacatecas Molecular Medicine Laboratory and the Public Health Laboratory in Zacatecas, Mexico, we compared the performance of the ARTIC V3 panel (218 primers) produced either by EDS or ordered from a commercial supplier. Libraries were generated from two synthetic SARS-CoV-2 RNA controls (10,000 genome copy equivalents per reaction), as well as five clinical samples (with RT-qPCR Cq values ranging between 18.5 and 30.9). Sequencing data for libraries generated with EDS and commercial primers showed no significant difference respect to coverage uniformity and variant calling (not shown).